Methylation Specific Bisulfite Seq Library Prep Kit
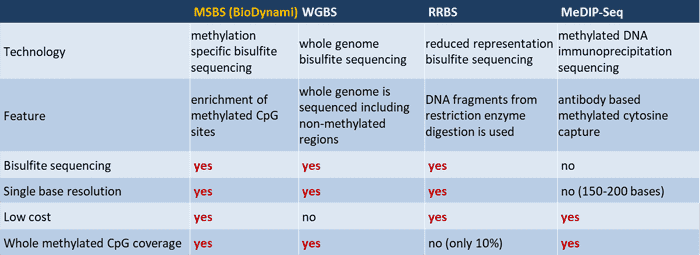
Bisulfite seq is a well know technology to detect DNA methylation and several technologies such as WGBS, RRBS, MeDIP-Seq, and MSBS are used for whole genome DNA methylation analysis. DNA methylation is important for regulation of cell development, differentiation and gene expression in molecular biology, genetics and epigenetics. Most methylated cytosines are found at CpG sites, and 70-80% of cytosines are methylated. The number of CpG sites in human genome is around 28 million, which is less than 1% of the genome compared with 4.4% expected.
Whole genome bisulfite sequencing (WGBS) is the most effective method of DNA methylation analysis. The only limitation is the sequencing cost is very high because the whole genome is sequenced including all the non-methylated regions.
Reduced Representation Bisulfite Sequencing (RRBS) is the reduced representation of a smaller fraction of the methylated CpG sites. RRBS combines restriction enzyme digestion and bisulfite sequencing, and enriches the sequencing for methylated CpG sites. It is an efficient technology for estimate the whole genome methylation patterns at the single base level. Although this allows a higher coverage depth and reduces the sequencing cost, the limitation is only 10% of the methylated CpG sites are covered.
Methylated DNA Immunoprecipitation Sequencing (MeDIP-Seq) is another whole genome enrichment technique used for selection of methylated DNA. Using antibodies against 5-methylcytosine, methylated DNA is enriched from whole genomic DNA via immunoprecipitation. 5-methylcytosine antibodies are incubated with fragmented genomic DNA and precipitated, followed by DNA purification and sequencing. There are several drawbacks of MeDIP-Seq: 1. Low resolution (150~200 bp) as opposed to the single base resolution; 2. Non-specific interaction due to antibody specificity and selectivity. 3. Bias towards hypermethylated regions.

Methylation Specific Bisulfite Seq Library Prep Kit Workflow
The Methylation Specific Bisulfite Seq (MSBS) Library Prep Kit (illumina platform) was developed for construction of NGS libraries for methylated CpG sites using bisulfite treated DNA (20 ng – 500 ng) as input. The kit enriches methylated CpG regions, thus significantly reduce the sequencing cost. The kit estimates the whole genome methylation patterns at the single base level since it is based on a bisulfite-seq technology.
It is known that bisulfite treatment of completed NGS libraries causes tremendous damage to the libraries. By using bisulfite treated DNA as input, the kit overcomes the significant library loss due to the bisulfite conversion. The kit contains a mixture of PCR polymerases that have high-fidelity amplification and uracil tolerance which is ideal for bisulfite treated DNA.
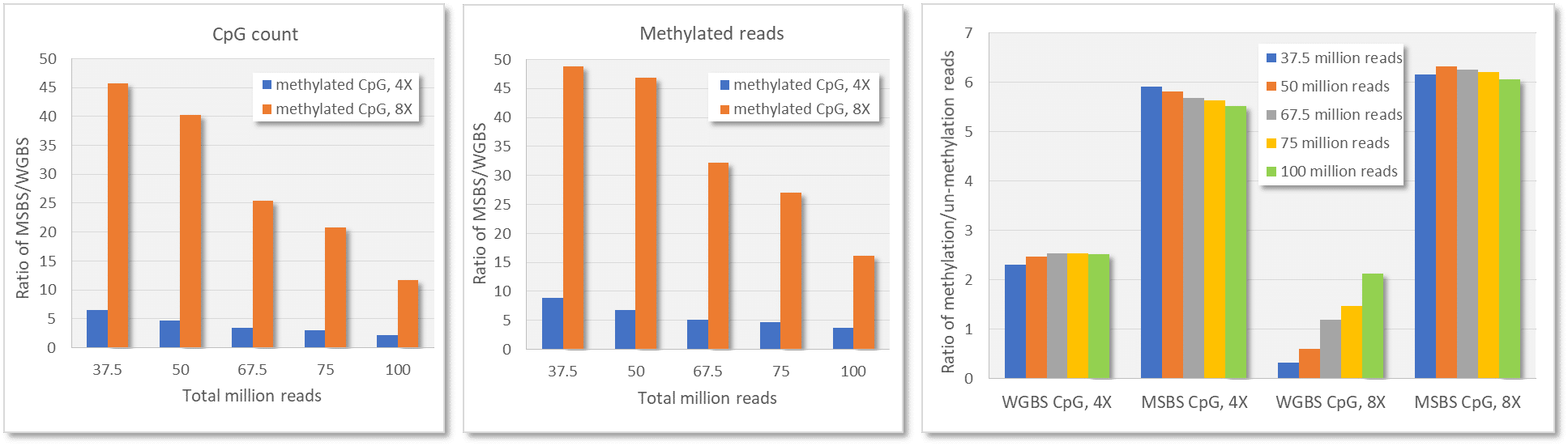
MSBS Library Prep Kit enriches CpG sites
Library multiplexing up to 96 samples is possible with the unique dual indexes. We have developed a 4-Base Difference Index System. The system can generate indexes with at least 4 bases different from others in the 8-base indexing region. The unique dual indexing primers identify sequencing errors such as index hopping, mis-assignment, and de-multiplexing errors. Index information can be downloaded here.
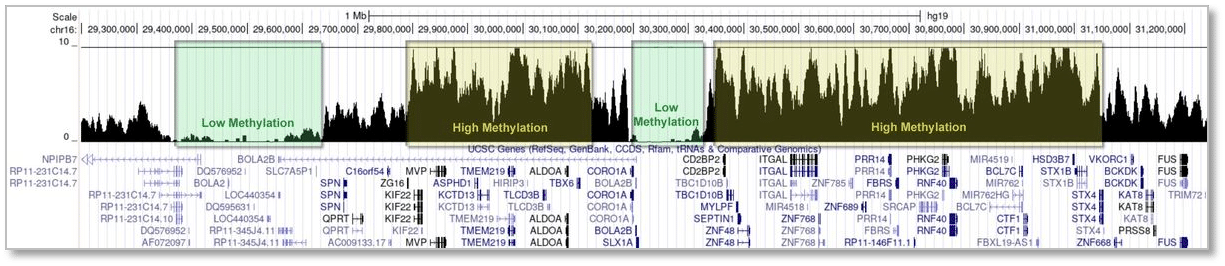
High methylation regions and low methylation regions in human genome
Features
- Enrichment of methylated CpG sites
- Single-base resolution
- Low cost for sequencing
- Fast
- Total time: 1.5 hours
- Hands-on time: 10 minutes
- Simple workflow
- Bisulfite treated DNA as input: From 20 ng to 500 ng
Protocol & MSDS Download
Product Category
Cat.# & Rxns
- Cat.# 30103A/30103F/30103S/30103L: 24/48/96/192 rxns
Related Products
- Bisulfite Sequencing Library Prep Kit
- Bisulfite Conversion Kit (Magnetic Beads)
- NGS DNA Fragmentation & Library Prep Kit
- Multiplexing UDI Primers (Illumina Platform)
- Multiplexing UDl Primers & Adaptors (Illumina Platform)
- NGS Library Quantification Standards With PCR Primers (Illumina platform)
- Library Size Selection Kit (250-350 bp, Magnetic Beads)
- Library Size Selection Kit (300-450 bp, Magnetic Beads)
- Library Size Selection Kit (450-750 bp, Magnetic Beads)
- Library Size Selection Kit (>5 kb, Magnetic Beads)
- Library Size Selection Kit (>10 kb, Magnetic Beads)
- DNA Normalization Magnetic Beads (NGS, PCR, gDNA)
- Magnetic Beads (DNA & RNA Purification)
Background info