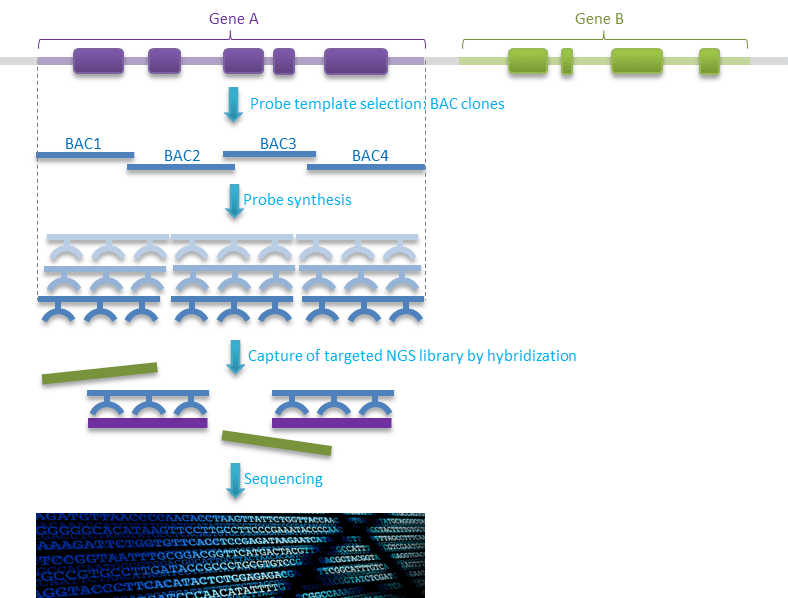
CATCH-Seq Technology
The proprietary CATCH-Seq targeted sequencing technology leverages genomic clones to generate probes corresponding to large, contiguous blocks of a genome. This is particularly useful when your research calls for sequencing coding and non-coding regions of a gene, including introns and flanking sequences. Sequencing large target regions including introns make it ideal for identification of structural variants and CNV’s.
CATCH-Seq target enrichment is also ideal for studies of DNA methylation, by performing targeted bisulfite sequencing after the target enrichment steps. CATCH-Seq target enrichment technology is compatible with any NGS library or platform. Best of all, since CATCH-Seq technology requires no oligonucleotide design and production costs, custom capture reagents are available at a fraction of the cost of other target enrichment methods.
Advantages of CATCH-Seq
Large target regions: CATCH-Seq capture probes can cover several hundred kilobases to several megabases of contiguous sequence and can cover exons, introns, 5’ regulatory regions, 3’ regulatory regions, and flanking regions.
Greater specificity with longer probes (200-300bp)
Better coverage of the targeted region including repeats
Significantly lower cost than oligo-based probes
Multiplexed capture of samples reduces costs
Featured Products
-
-
- NGS DNA Library Prep Kit
- NGS DNA Library Prep Customization Kit
- PCR-free NGS DNA Library Prep Kit
- NGS Low Input DNA Library Prep Kit
- ChIP-Seq Library Prep Kit
- NGS Cell-free DNA Library Prep Kit
- NGS FFPE DNA Library Prep Kit
- Bisulfite Sequencing Library Prep Kit
- Methylation Specific Bisulfite-Seq Library Prep Kit
- NGS Single Stranded DNA Library Prep Kit
- NGS Ancient DNA Library Prep Kit
- NGS DNA Fragmentation & Library Prep Kit
- RNA Seq Library Prep Kit
-
Related Products
-
-
- MHC Capture Kit
- MHC Library Prep & Capture Kit
- MHC Class I Capture Kit
- MHC Class I Library Prep & Capture Kit
- MHC Class II Capture Kit
- MHC Class II Library Prep & Capture Kit
- MHC Class III Capture Kit
- MHC Class III Library Prep & Capture Kit
- MHC Core Capture Kit
- MHC Core Library Prep & Capture Kit
- LRC/KIR Capture Kit
- LRC/KIR Library Prep & Capture Kit
-
Examples of CATCH-Seq technology
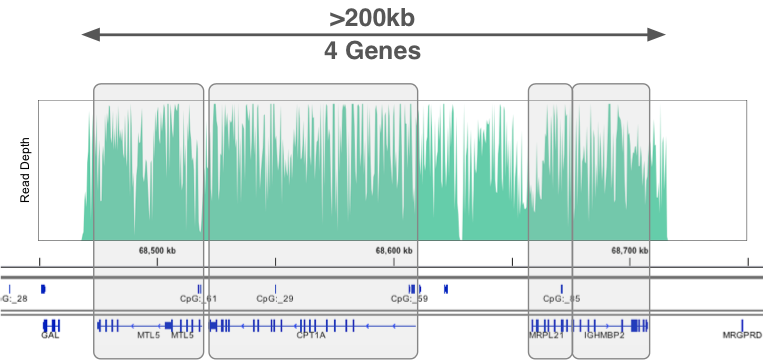
Targeted sequencing of four genes (>200 kb region)
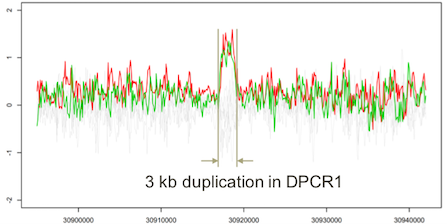
Targeted sequencing of DPCR1: The detection of 3 kb duplication region
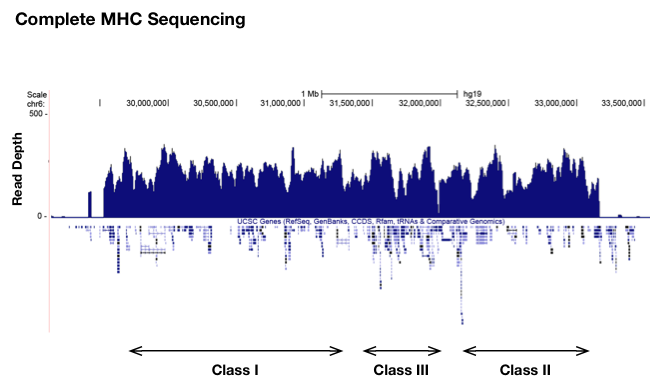
Targeted sequencing of MHC (~3.8 MB)
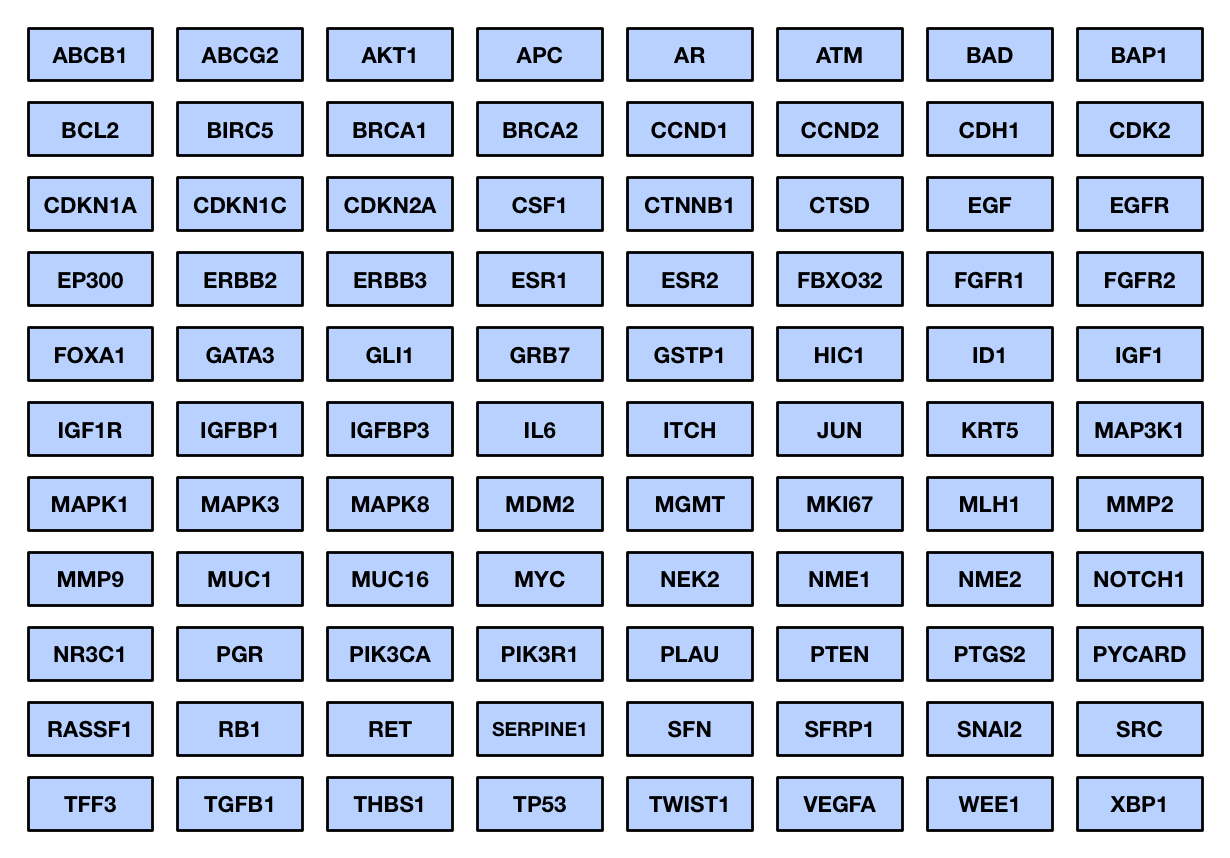
Breast Cancer Panel: up to 88 genes related to breast cancer, including all coding and non-coding regions
Service and Product with CATCH-Seq Technology
Targeted Sequencing Service
MHC sequencing: Capture and sequence the human MHC locus including exons, introns, and flanking sequences.
LRC/KIR sequencing: Capture and sequence the human KIR locus including exons, introns, and flanking sequences.
Breast cancer panel: Capture and sequence coding and non-coding regions of up to 88 human genes related to breast cancer.
Custom CATCH-Seq targeted sequencing: Capture and sequence the customized large target regions of 200 kb to >10 Mb.
Targeted Sequencing Product
MHC sequencing: Capture and sequence the human MHC locus including exons, introns, and flanking sequences.
LRC/KIR sequencing: Capture and sequence ~X Mb of the human KIR locus including exons, introns, and flanking sequences.
Breast cancer panel: Capture and sequence coding and non-coding regions of 88 breast cancer related genes.
Publications using CATCH-Seq
Targeted sequencing of large genomic regions with CATCH-Seq.
PLoS One. 2014 Oct 30; 9(10):e111756. PMID: 25357200
Epigenome-wide association study of fasting blood lipids in the Genetics of Lipid-lowering Drugs and Diet Network study.
Circulation. 2014 Aug 12; 130(7):565-72. PMID: 24920721
Regulation of DNA methylation dictates CD4 expression during the development of helper and cytotoxic T cell lineages.
Nat Immunol. 2015 Jul; 16(7):746-54. PMID: 26030024
Recurrent evolution of melanism in South American felids.
PLoS Genet. 2015 Feb 19;11(2):e1004892. doi: 10.1371/journal.pgen.1004892. PMID: 25695801
A deletion at ADAMTS9-MAGI1 locus is associated with psoriatic arthritis risk.
Ann Rheum Dis. 2015 Oct;74(10):1875-81. doi: 10.1136/annrheumdis-2014-207190. PMID: 25990289
dCATCH-Seq: improved sequencing of large continuous genomic targets with double-hybridization.
BMC Genomics 2017:18, 811. DOI: https://doi.org/10.1186/s12864-017-4159-7
Stage-specific epigenetic regulation of CD4 expression by coordinated enhancer elements during T cell development.
Nat Commun. 2018 Sep 5;9(1):3594. doi: 10.1038/s41467-018-05834-w. PMID: 30185805
Whole Blood Targeted Bisulfite Sequencing Validates Differential Methylation in C6ORF10 gene of Patients with Rheumatoid Arthritis.
J Rheumatology. 2019 Nov; jrheum.190376; DOI: 10.3899/jrheum.190376
TET proteins regulate T cell and iNKT cell lineage specification in a TET2 catalytic dependent manner.
Front Immunol. 2022 Aug 5;13:940995. doi: 10.3389/fimmu.2022.940995. PMID: 35990681; PMCID: PMC9389146.
CD4 expression in effector T cells depends on DNA demethylation over a developmentally established stimulus-responsive element.
Nat Commun. 2022 Mar 18;13(1):1477. doi: 10.1038/s41467-022-28914-4. PMID: 35304452