Library Size Selection Kit (for Short-read Sequencing, Magnetic Beads)
The size selection ranges ideal for short-read sequencing:
-
- Cat.# 20104: 250-350 bp (for illumina PE100 sequencing)
- Cat.# 20105: 300-450 bp (for illumina PE150 sequencing)
- Cat.# 20106: 450-750 bp (for illumina PE300 sequencing)
The Library Size Selection Kit (for Short-read Sequencing, Magnetic Beads) can be used for library size selection for short-read sequencing with double-sided clean-ups. DNA size selection is preferred after NGS library prep in most of the cases.
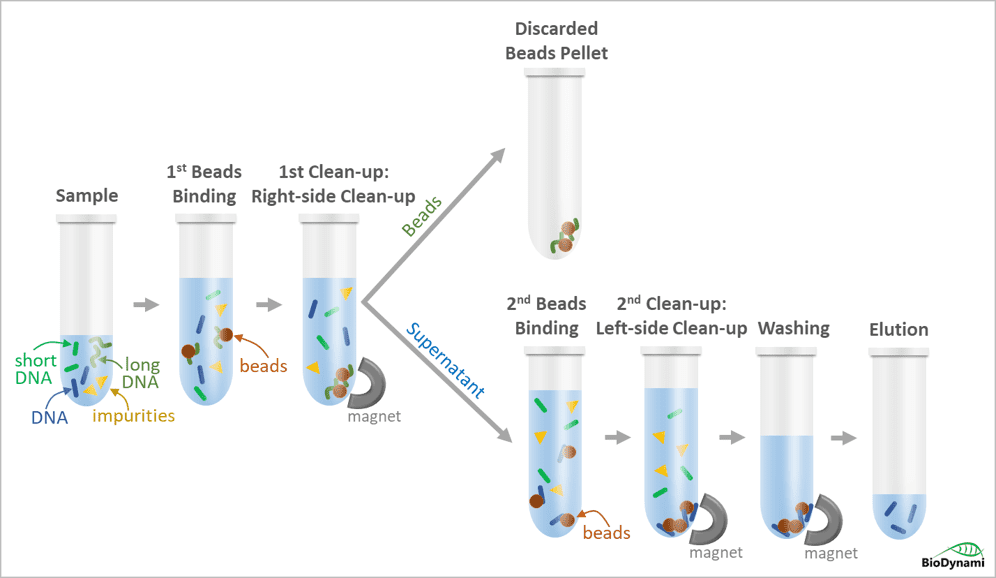
DNA size selection with dual clean-ups.
Library size selection is an enrichment of a specific range of library sizes for NGS library preparations. The NGS library preparation is related to the quality of the sequencing data. Precise NGS library size selection can increase sequencing efficiency, improve data quality, and reduce costs.
Short-read sequencing uses DNA libraries that contain small insert DNA fragments of similar sizes, usually several hundred base pairs. The sequencing efficiency can be improved if the DNA size selection is in the right range. Cat.# 20104S and 20104L are the best kits for NGS library size selection of illumina paired-end 100 (PE100) sequencing with 100-200 bp library inserts; Cat.# 20105S and 20105L are the best kits for NGS library size selection of illumina paired-end 150 (PE150) sequencing with 150-300 bp library inserts; and Cat.# 20106S and 20106L are the best kits for NGS library size selection of illumina paired-end 300 (PE300) sequencing with 300-600 bp library inserts.

Gel images of different ranges of size selection. Sheared human genomic DNA was used as input.
The magnetic beads, or SPRI (Solid Phase Reversible Immobilization) beads, is well used for the purification of DNA due to their reversible DNA binding. The NGS library can be size-selected by the magnetic beads or SPRI beads. The properties of the magnetic beads can be changed for a specific range of DNA binding. The contaminants and other unwanted components in the libraries can also be removed during size selection.
Specific ranges of NGS libraries can be selected using magnetic beads with different buffer compositions. The first DNA-beads binding step, also called the right-side clean-up, removes large NGS library fragments. The large NGS library fragments that bind to the beads are discarded with the beads pellet. The desired NGS library fragments in the supernatant are transferred to a new well, and new beads are added to the supernatant for the second beads-DNA binding, also called the left-side clean-up. After the rinsing step, the NGS library fragments with the dual selection are eluted in water or an appropriate buffer. The magnetic beads method has great advantages over time-consuming column purification and tedious gel-based purification.
Features
- High specificity and high recovery of size selection
- 3 ranges for short-read sequencing size selection
- illumina PE100 sequencing: 250-350 bp
- illumina PE150 sequencing: 300-450 bp
- illumina PE300 sequencing: 450-750 bp
- Rapid protocol: only 20 minutes
- Simple workflow
- No columns required
- No gel purification required
- No centrifugation required
- Efficient removal of contaminants and unwanted components
Protocol & MSDS Download
Product Category
Cat.# & Rxns
- Cat.# 20104S/20104L (illumina PE100 sequencing, 250-350 bp): 24/96 rxns
- Cat.# 20105S/20105L (illumina PE150 sequencing, 300-450 bp): 24/96 rxns
- Cat.# 20106S/20106L (illumina PE300 sequencing, 450-750 bp): 24/96 rxns
Related Products
- DNA Size Selection Kit (50-100 bp, Magnetic Beads)
- DNA Size Selection Kit (100-200 bp, Magnetic Beads)
- DNA Size Selection Kit (200-500 bp, Magnetic Beads)
- DNA Size Selection Kit (250-350 bp, Magnetic Beads)
- DNA Size Selection Kit (300-450 bp, Magnetic Beads)
- DNA Size Selection Kit (450-750 bp, Magnetic Beads)
- DNA Size Selection Kit (500-1000 bp, Magnetic Beads)
- DNA Size Selection Kit (1-3 kb, Magnetic Beads)
- DNA Size Selection Kit (1-5 kb, Magnetic Beads)
- DNA Size Selection Kit (>5 kb, Magnetic Beads)
- DNA Size Selection Kit (>10 kb, Magnetic Beads)
Background info
