
SPRI Beads Technology: Unlocking the Power of a Robust Lab Workhorse
What are SPRI beads?
SPRI stands for Solid Phase Reversible Immobilization, a method that uses paramagnetic beads coated with carboxylate (or similar) groups to bind nucleic acids (DNA/RNA) in the presence of polyethylene glycol (PEG) and salt. SPRI beads have become the undisputed backbone of genomics and molecular biology for purification of DNA and RNA.
SPRI beads are well-used for fast, efficient purification and size selection of DNA/RNA, working by binding nucleic acids in specific buffer conditions and being separated with a magnet, making workflows simpler and more automated than traditional methods like centrifuge and spin columns.
What SPRI beads are made of?
- Polystyrene core
- Paramagnetic layer (iron oxide)
- Carboxylate-coated surface or similar
- Suspended in a buffer containing
-
- PEG (polyethylene glycol)
- Salt (usually sodium chloride)
-
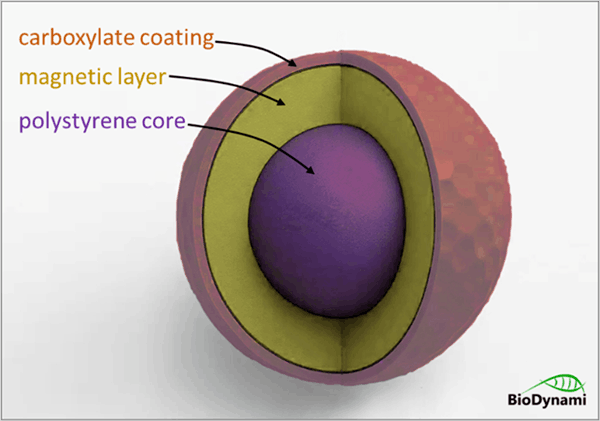
SPRI beads typically consist of a polystyrene core with a magnetic layer (magnetite), and a specialized surface coating, most commonly Carboxylate groups (-COOH) for reversible, size-selective nucleic acid binding. These Carboxylate-coated magnetic particles are essential for NGS library cleanup, DNA/RNA purification, and DNA/RNA extraction by enabling, with PEG and salt, the selective binding and purification of nucleic acids.
Nucleic Acid Purification Beads
DNA Purification
- Magnetic Beads (DNA & RNA Purification)
- Magnetic Beads (microRNA & Oligo Purification)
- Magnetic Beads (Short Oligo Purification)
- Magnetic Beads (PCR Purification)
- DNA Concentrator (Magnetic Beads)
- cfDNA Purification Kit (Magnetic Beads)
- Plasmid Purification Magnetic Beads (RNA Depletion)
- RNA Contamination Removal Magnetic Beads (gDNA Purification)
- RNase-free RNA Removal (Magnetic Beads)
RNA Purification
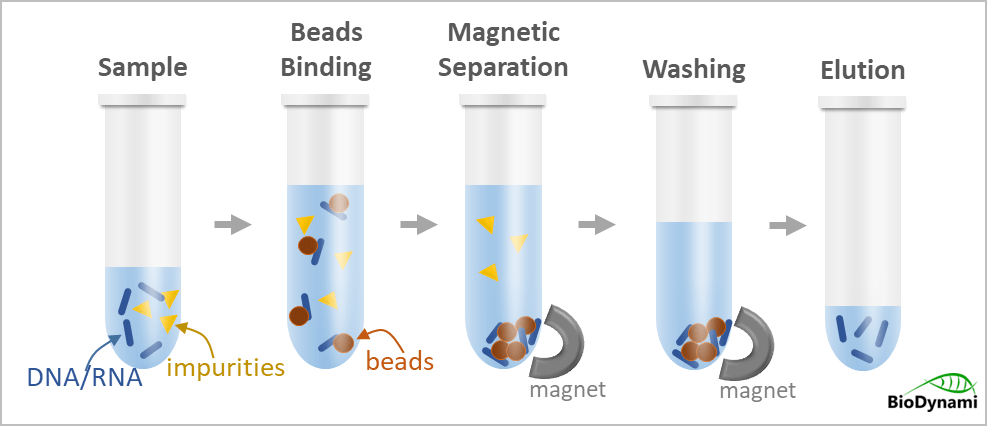
What is the typical workflow for a SPRI beads cleanup?
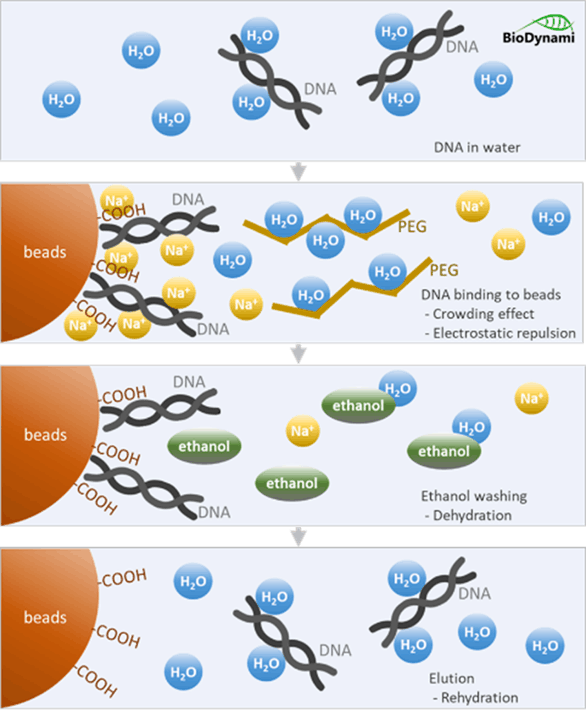
- Binding: SPRI beads have a magnetic core (magnetite) coated with carboxyl groups that bind DNA/RNA. A special buffer (with PEG and salt) creates crowding, forcing nucleic acids and bead surfaces to come close enough to bind.
- Magnetic separation: A magnet pulls the beads (with bound DNA/RNA) to the side of the tube, allowing unwanted solution (salts, enzymes, short fragments) to be discarded.
- Washing: Ethanol washes remove remaining impurities.
- Elution: An aqueous buffer releases the purified DNA/RNA from the beads.
Why SPRI beads can bind to DNA?
Main factors related to DNA binding
1. Carboxyl-coated surface: The paramagnetic beads (polystyrene core with a magnetite layer) are coated with carboxyl groups (-COOH), they don’t retain magnetism once the field is removed.
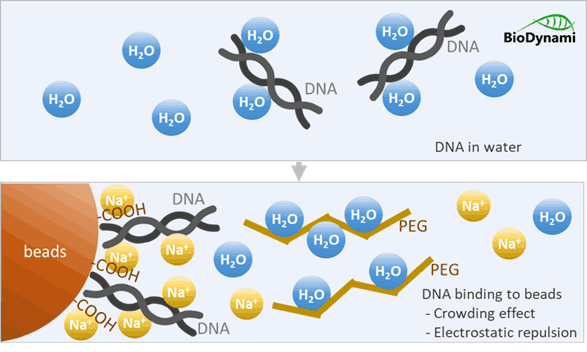
2.PEG causes molecular crowding: Acting as a crowding agent. PEG is a large, water-loving polymer that takes up space in solution and reduces the amount of water available to solvate DNA. PEG dehydrates the DNA, reducing its solubility and forcing it to precipitate out of the solution toward the carboxyl groups on the bead surface.
This creates a crowding effect:
-
- No water molecules for DNA: The highly hydrophilic PEG molecules aggressively attract and bind water molecules, effectively “stealing” the water away from other solutes, including DNA.
- DNA forced out of solution: DNA is a charged, hydrophilic molecule normally surrounded by a shell of water. As PEG sequesters water, this shell is disrupted. The DNA is forced out of solution.
3.Salt shields negative charges
Both DNA and the bead surface are negatively charged. Normally they would repel each other, but in the presence of Salt (Na⁺ ions):
-
- Neutralizes negative charges
- Reduces electrostatic repulsion
- Allows DNA to approach the bead surface
This screens the electrostatic repulsion, allowing the now less-charged DNA to come very close to the beads.
DNA binding is based on dehydrated environment
This is not sequence-specific and not chemical bonding. In this water-depleted, crowded environment, the hydrophobic components of DNA and the hydrophobic regions on the bead surface interact preferentially. The DNA precipitates out of the bulk solution and coats the bead surface. At this stage, the DNA is not covalently attached; it’s physically adsorbed due to a combination of hydrophobic interactions and van der Waals forces in a crowded, low-water setting.
In essence, SPRI beads don’t magically bind to DNA. They provide a hydrophobic surface in a chemically controlled environment that first induces DNA precipitation onto itself, and then allows for clean re-solubilization when that environment is changed. This reversible, solid-phase immobilization is what gives the technology its name and its power.
What SPRI beads binding is NOT?
- Not sequence-specific
- Not affinity binding
- Not covalent chemistry
- Not magnetic attraction to DNA
The magnet only moves the beads, not DNA.
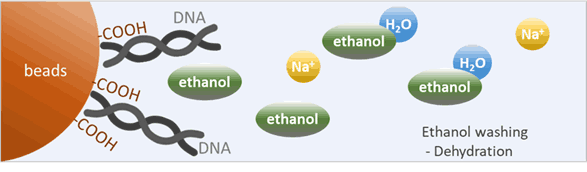
What is the purpose of washing beads with ethanol?
After DNA-binding, the supernatant is removed, and 80% ethanol is added as a wash. Ethanol is even more dehydrating than PEG. It further reduces the water available, strengthening the hydrophobic interactions.
The ethanol wash could:
- Ensuring the DNA remains tightly bound to the beads. This could increase the DNA yield of the elution step.
- Remove impurities and residual salts/PEG from the bead pellet.
How elution works (Reversing the conditions; the “R” in SPRI)?
Reversible binding: The binding is not permanent. The DNA can be eluted from the beads using a low-salt buffe or water.
When you remove PEG/salt and add elution buffer:
- DNA rehydrates
- Electrostatic repulsion returns
- DNA releases back into solution
To release the DNA:
- The ethanol is completely removed, and the bead pellet is briefly air-dried.
- A low-salt, aqueous buffer is added.
- Key Reversal: The elution buffer has no PEG and very low salt. Water molecules are now freely available to rehydrate the DNA.
- The DNA dissolves back into the aqueous solution, leaving the beads behind.
Advantages
- Size selectivity: The precise ratio of bead/sample determines the binding cutoff, allowing isolation of fragments above a specific size (for example: a 1x ratio removes short primers, while a 0.6x ratio selects for longer DNA fragments). You can enrich for specific DNA fragment sizes, useful for NGS library prep and amplicon cleanups.
- High throughput & automation: Compatibility with liquid handlers has made them essential for high-throughput. Perfectly suited for 96-well plates and liquid handling robots, enabling scalable, consistent processing with minimal hands-on time.
- Cost-effectiveness: Compared to traditional column-based kits, bulk SPRI reagents offer a dramatically lower cost per sample without sacrificing yield or purity for standard applications.
- Flexibility & versatility: A single reagent can be used for a wide array of applications: double-stranded DNA, single-stranded DNA, RNA, end-repair, A-tailing, ligation, and adapter clean-up across a wide range of input masses (nanograms to micrograms).
- Excellent recovery & purity: Modern formulations offer high recovery rates (>80-90%) for most fragments and effectively remove salts, enzymes, nucleotides, and other common contaminants.
- Simple workflow: Magnetic separation replaces centrifugation, making protocols faster and easier.
- Low contamination risk: You can avoid toxic chaotropic salts or organic solvents used in traditional methods, reducing contamination and safety issues.
Limitations
- Technique sensitivity:
-
- Beads settle over time and must be fully resuspended before use. Incorrect ratios or incomplete resuspension can lead to poor yields.
- Manual pipetting of bead mixtures, especially during the alcohol wash steps, remains a source of variability. Incomplete removal of ethanol is the most common cause of downstream enzymatic inhibition.
-
- Fragment size limitations:
-
- Small fragments: Standard SPRI beads struggle to recover fragments <100 bp efficiently, meaning they aren’t ideal for very short oligos unless specialized beads are used. Recent progress has made it possible to recover tRNA (around 70-90 nt), miRNA (around 21 nt), and short oligos (as short as 6 nt).
- Less optimal for ultra-high-molecular-weight DNA: Some users report lower yields or extra handling for very large genomic DNA (>10 kb). Adjustments in bead volumes and conditions are often needed.
-
- Beads-drying issues: Over-dried beads after wash steps can dramatically reduce recovery. Many labs share anecdotes about difficulties with drying and elution.
- Size selection resolution: While excellent for removing very small fragments, the cutoff is not a precise “hard wall.” There is a distribution, meaning some fragments below the target size may be retained and some above may be lost.
- Beads loss: Over-vigorous pipetting or mis-handling can lead to bead loss, reducing yield. Consistent, gentle mixing is key.
Applications
Since their widespread adoption, they have become the foundational tool for genomics and molecular biology, largely displacing older silica-column methods in high-throughput and automated workflows. Recent developments have expanded the beads applications, including small DNA/RNA fragments recovery, separation of small cfDNA fraction from large genomic DNA, DNA/library normalization, etc.
- NGS library preparation
- dsDNA, ssDNA, and RNA clean-up
- PCR Clean-up
- Genomic DNA Clean-up with RNA depletion
- Plasmid Clean-up with RNA depletion
- cfDNA clean-up with removal of gDNA fraction
- miRNA clean-up
- tRNA clean-up
- Oligo clean-up (as small as 6 nt)
- Size selection for DNA fragments and NGS libraries
- DNA normalization
- DNA Concentrator
- RNase-free RNA removal (for RNA labs)
Trends
SPRI beads are standard now in modern molecular biology workflows thanks to their simplicity, efficiency, and scalability. SPRI is no longer a single product but a technology platform. Multiple vendors offer SPRI beads with proprietary surface chemistries and formulations.
- Enhanced Beads: Formulations designed for superior recovery of very short (<100 bp) or very long (>1000 bp) fragments, or for ultralow-input samples (e.g., single-cell genomics).
- Automation-Optimized: Beads with adjusted viscosity and magnetic properties for specific liquid handlers.
- Sustainability: Some vendors offer concentrated stocks to reduce plastic packaging and shipping weight.
- Rigorous QC: Leading suppliers provide detailed quality control data, including fragment analysis recovery profiles, ensuring lot-to-lot consistency critical for reproducible research and diagnostics.
Conclusion
SPRI beads are a transformative technology that has enabled the scale and efficiency of genomics and molecular biology. Their simplicity, versatility, and cost-effectiveness make them an indispensable tool in academic, clinical, and industrial labs. Their continued evolution with specialized formulations ensures they will remain a core reagent in the field of molecular biology and genomics.