DNA Fragmentation Enzyme
The DNA fragmentation enzyme was developed for random fragmentation of genomic DNA (EDTA-free DNA or DNA resuspended in TE buffer) in a fast and simple way. The DNA fragments contain 5´-phosphates and 3´-hydroxyl groups.
Enzyme-based fragmentation is very simple and effective comparing to the mechanical shearing methods. There are several advantages of enzymatic DNA fragmentation, include easy handling of many samples at the same time, and less loss of samples. Moreover, it does not require expensive capital expense for the equipment.
The resulting DNA fragment size is inversely correlated with the incubation time of step 1 at 20°C. The fragmented DNA can be used for applications such as Next-Generation Sequencing (NGS). Our Fragmentation Enzyme does not generate detectable sequencing bias. Sequence coverage is also consistent between enzymatic and mechanical fragmentation.
Cat.# 40061 | DNA Fragmentation Enzyme Mix |
Cat.# 40062 | DNA Fragmentation & A-tailing Enzyme Mix |
Cat.# 40063 | DNA Fragmentation & Blunting Enzyme Mix |
DNA Fragmentation Enzyme Mix (Cat. # 40061)
The enzyme mix was developed for enzymatic fragmentation of genomic DNA with random ends in a single step. The fragmented DNA can be used for applications such as Next-Generation Sequencing (NGS).
Enzyme Workflow
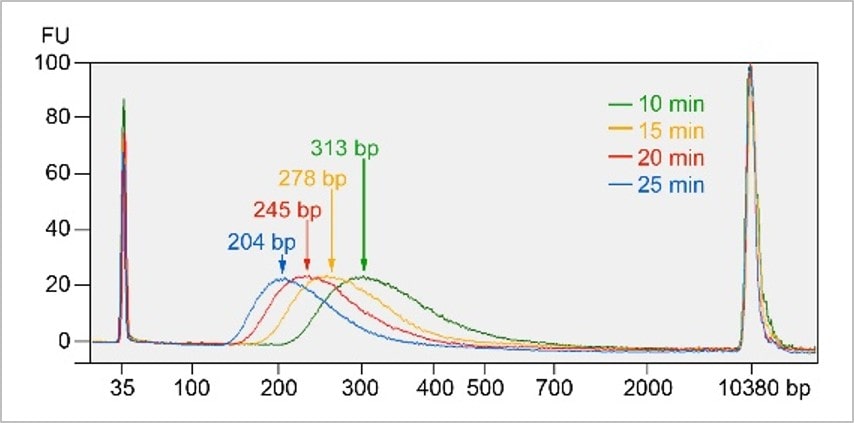
DNA fragment size is inversely correlated with the incubation time of step 1 at 20°C.
DNA Fragmentation & A-tailing Enzyme Mix (Cat. # 40062)
The enzyme mix can be used for enzymatic fragmentation of genomic DNA and addition of A-tailing at 3’-ends of DNA in a single step. 3’-ends of DNA with A-tailing can be used in many downstream applications in the field of molecular biology such as next generation sequencing, PCR cloning, and TA-ligation etc.
DNA Fragmentation & A-tailing Workflow
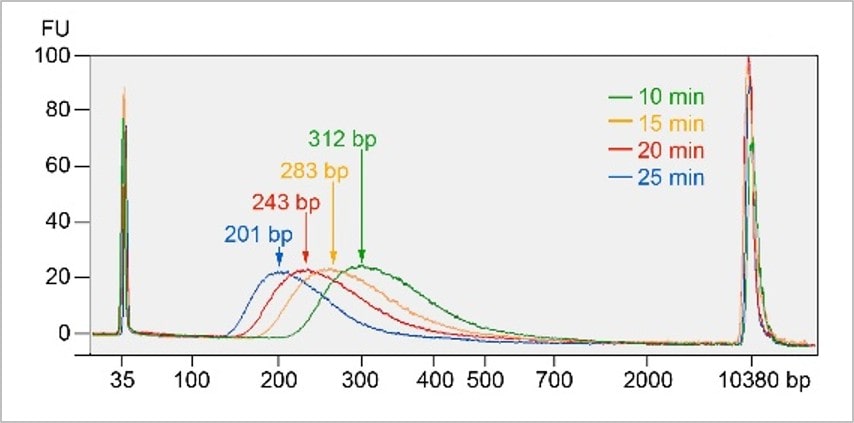
DNA fragment size is inversely correlated with the incubation time of step 1 at 20°C.
DNA Fragmentation & Blunting Enzyme Mix (Cat. # 40063)
The enzyme mix was developed to generate random shearing of genomic DNA with blunt ends in a single step. The fragmented DNA can be used for applications such as Next-Generation Sequencing (NGS), blunt end ligation and PCR cloning.
DNA Fragmentation & Blunting Workflow
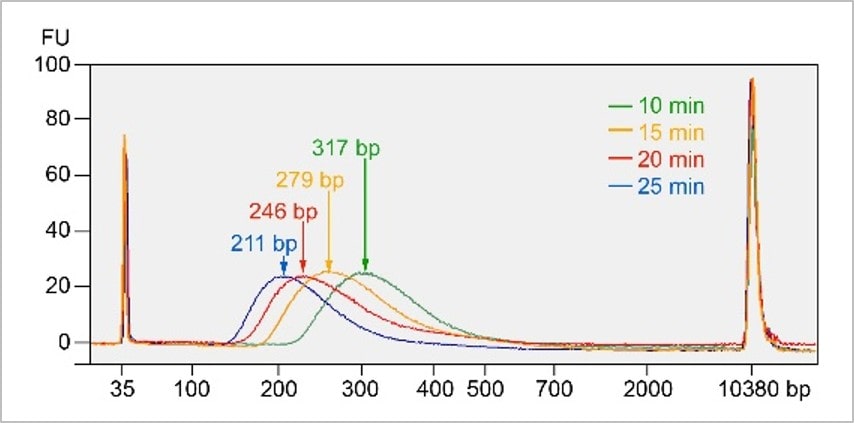
DNA fragment size is inversely correlated with the incubation time of step 1 at 20°C.
Features
- Fast enzymatic fragmentation: 30-45 minutes
- Simple Procedure: 3 minutes of hands-on time
- Works with both EDTA-free DNA and DNA resuspended in TE buffer
- Ideal for downstream reactions: NGS library prep, PCR cloning, TA-ligation, and blunt end ligation etc.
Specifications
DNA Fragmentation Enzyme Mix library | DNA Fragmentation & Blunting Enzyme Mix | DNA Fragmentation & A-tailing Enzyme Mix | |
Cat.# | 40061 | 40063 | 40062 |
Protocol time | 30-45 min | 30-45 min | 30-45 min |
Hands-on time | 3 min | 3 min | 3 min |
Compatibility with EDTA-free DNA | |||
Compatibility with DNA in TE buffer | |||
One-step reaction | |||
Ends of DNA fragments | Random | Blunt | 3' A-tailed |
Applications | NGS library prep | NGS library prep; PCR cloning; blunt end ligation | NGS library prep; PCR cloning; TA ligation |
Protocol & MSDS Download
Cat.# & Rxns
- Cat.# 40061S/40061L: 50/200 rxns
- Cat.# 40062S/40062L: 50/200 rxns
- Cat.# 40063S/40063L: 50/200 rxns
Related Products
- Blunt/TA Ligation Master Mix
- Rapid Ligation Master Mix
- Sticky End Ligation Master Mix
- End Repair/dA-tailing Master Mix
- NGS Ligation Enzyme
- Proteinase K
- Lysozyme
- RNase A
Ref